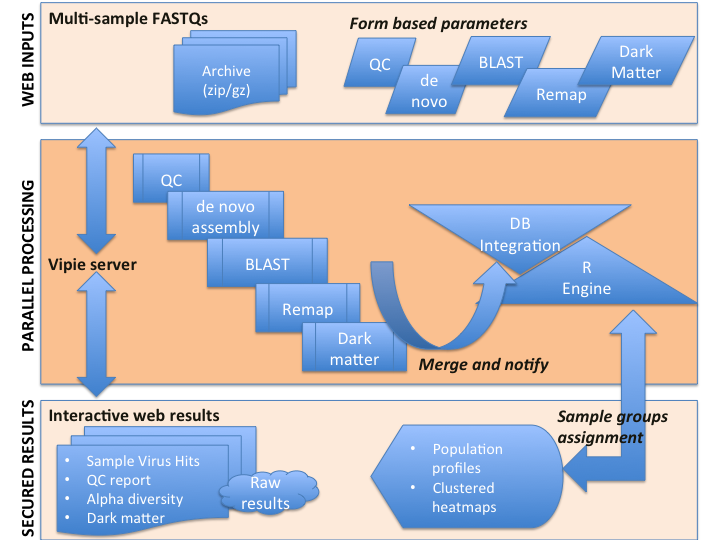
Name Email Password Institute (include URL) Country
Afghanistan
Åland Islands
Albania
Algeria
American Samoa
Andorra
Angola
Anguilla
Antarctica
Antigua and Barbuda
Argentina
Armenia
Aruba
Australia
Austria
Azerbaijan
Bahamas
Bahrain
Bangladesh
Barbados
Belarus
Belgium
Belize
Benin
Bermuda
Bhutan
Bolivia, Plurinational State of
Bonaire, Sint Eustatius and Saba
Bosnia and Herzegovina
Botswana
Bouvet Island
Brazil
British Indian Ocean Territory
Brunei Darussalam
Bulgaria
Burkina Faso
Burundi
Cambodia
Cameroon
Canada
Cape Verde
Cayman Islands
Central African Republic
Chad
Chile
China
Christmas Island
Cocos (Keeling) Islands
Colombia
Comoros
Congo
Congo, the Democratic Republic of the
Cook Islands
Costa Rica
Côte d'Ivoire
Croatia
Cuba
Curaçao
Cyprus
Czech Republic
Denmark
Djibouti
Dominica
Dominican Republic
Ecuador
Egypt
El Salvador
Equatorial Guinea
Eritrea
Estonia
Ethiopia
Falkland Islands (Malvinas)
Faroe Islands
Fiji
Finland
France
French Guiana
French Polynesia
French Southern Territories
Gabon
Gambia
Georgia
Germany
Ghana
Gibraltar
Greece
Greenland
Grenada
Guadeloupe
Guam
Guatemala
Guernsey
Guinea
Guinea-Bissau
Guyana
Haiti
Heard Island and McDonald Islands
Holy See (Vatican City State)
Honduras
Hong Kong
Hungary
Iceland
India
Indonesia
Iran, Islamic Republic of
Iraq
Ireland
Isle of Man
Israel
Italy
Jamaica
Japan
Jersey
Jordan
Kazakhstan
Kenya
Kiribati
Korea, Democratic People's Republic of
Korea, Republic of
Kuwait
Kyrgyzstan
Lao People's Democratic Republic
Latvia
Lebanon
Lesotho
Liberia
Libya
Liechtenstein
Lithuania
Luxembourg
Macao
Macedonia, the former Yugoslav Republic of
Madagascar
Malawi
Malaysia
Maldives
Mali
Malta
Marshall Islands
Martinique
Mauritania
Mauritius
Mayotte
Mexico
Micronesia, Federated States of
Moldova, Republic of
Monaco
Mongolia
Montenegro
Montserrat
Morocco
Mozambique
Myanmar
Namibia
Nauru
Nepal
Netherlands
New Caledonia
New Zealand
Nicaragua
Niger
Nigeria
Niue
Norfolk Island
Northern Mariana Islands
Norway
Oman
Pakistan
Palau
Palestinian Territory, Occupied
Panama
Papua New Guinea
Paraguay
Peru
Philippines
Pitcairn
Poland
Portugal
Puerto Rico
Qatar
Réunion
Romania
Russian Federation
Rwanda
Saint Barthélemy
Saint Helena, Ascension and Tristan da Cunha
Saint Kitts and Nevis
Saint Lucia
Saint Martin (French part)
Saint Pierre and Miquelon
Saint Vincent and the Grenadines
Samoa
San Marino
Sao Tome and Principe
Saudi Arabia
Senegal
Serbia
Seychelles
Sierra Leone
Singapore
Sint Maarten (Dutch part)
Slovakia
Slovenia
Solomon Islands
Somalia
South Africa
South Georgia and the South Sandwich Islands
South Sudan
Spain
Sri Lanka
Sudan
Suriname
Svalbard and Jan Mayen
Swaziland
Sweden
Switzerland
Syrian Arab Republic
Taiwan, Province of China
Tajikistan
Tanzania, United Republic of
Thailand
Timor-Leste
Togo
Tokelau
Tonga
Trinidad and Tobago
Tunisia
Turkey
Turkmenistan
Turks and Caicos Islands
Tuvalu
Uganda
Ukraine
United Arab Emirates
United Kingdom
United States
United States Minor Outlying Islands
Uruguay
Uzbekistan
Vanuatu
Venezuela, Bolivarian Republic of
Viet Nam
Virgin Islands, British
Virgin Islands, U.S.
Wallis and Futuna
Western Sahara
Yemen
Zambia
Zimbabwe
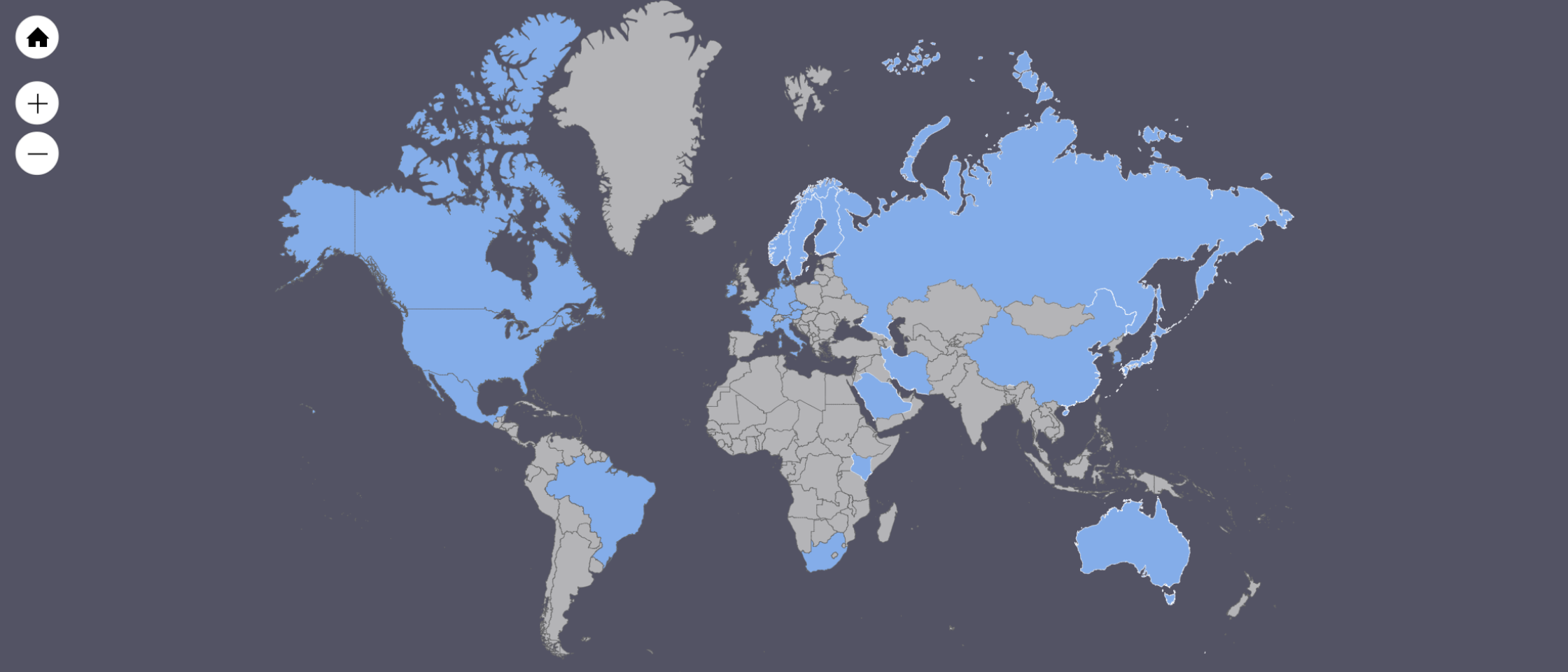
Researchers from 20+ countries (shown below) have registered for Vipie